Gene Updater: a web tool that autocorrects and updates for Excel misidentified gene names
A Streamlit webtool that converts date terms into updated gene symbols
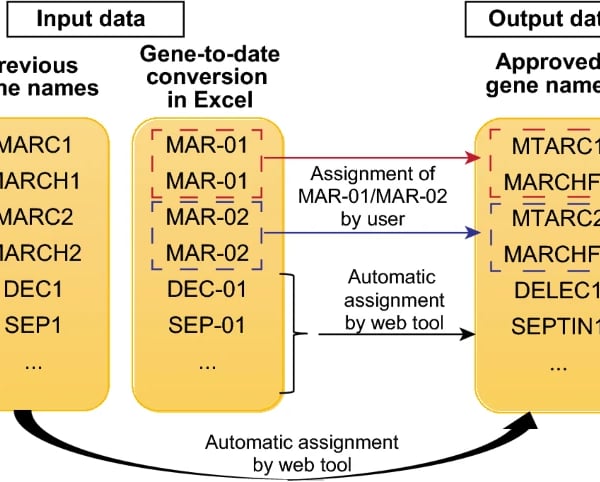
When gene expression data is opened in Excel, some of the gene symbols will be autoconverted into date terms. This can affect downstream pathway analysis as many of these databases rely on gene symbols to detect for pathway enrichment. To circumvent this limitation, we used Streamlit to create a web tool called Gene Updater that allows users to convert the date terms to the updated gene terms recommended by HUGO, which are more resilient to autoconversion by Excel. The webpage is hosted at: https://share.streamlit.io/kuanrongchan/date-to-gene-converter/main/date_gene_tool.py.
Users may visit our GitHub address to download the date_gene_tool.py file to understand the Python codes that allows construction of such a web tool. To run the web tool locally, users can download all the required Python packages and files at the designated GitHub address or at https://zenodo.org/record/6845701#.YtvCYiX0rDs.
To illustrate the importance and utility of this web tool, we downloaded the supplementary files from several of the top journals in the last month. This pursuit yielded 28/81 tables with date terms in their data files, of which 6 of them had no gene description columns, which can make the interpretation of these data files challenging. Fortunately, most of these errors can simply be corrected by Gene Updater, ensuring consistency in data sharing and data communication.
For more information of the web tool, users can read the scientific article currently published in Scientific Reports. This work is done in Duke-NUS Medical School, and special thanks to Clara Koh and Justin Ooi for creating and designing the web tool.
